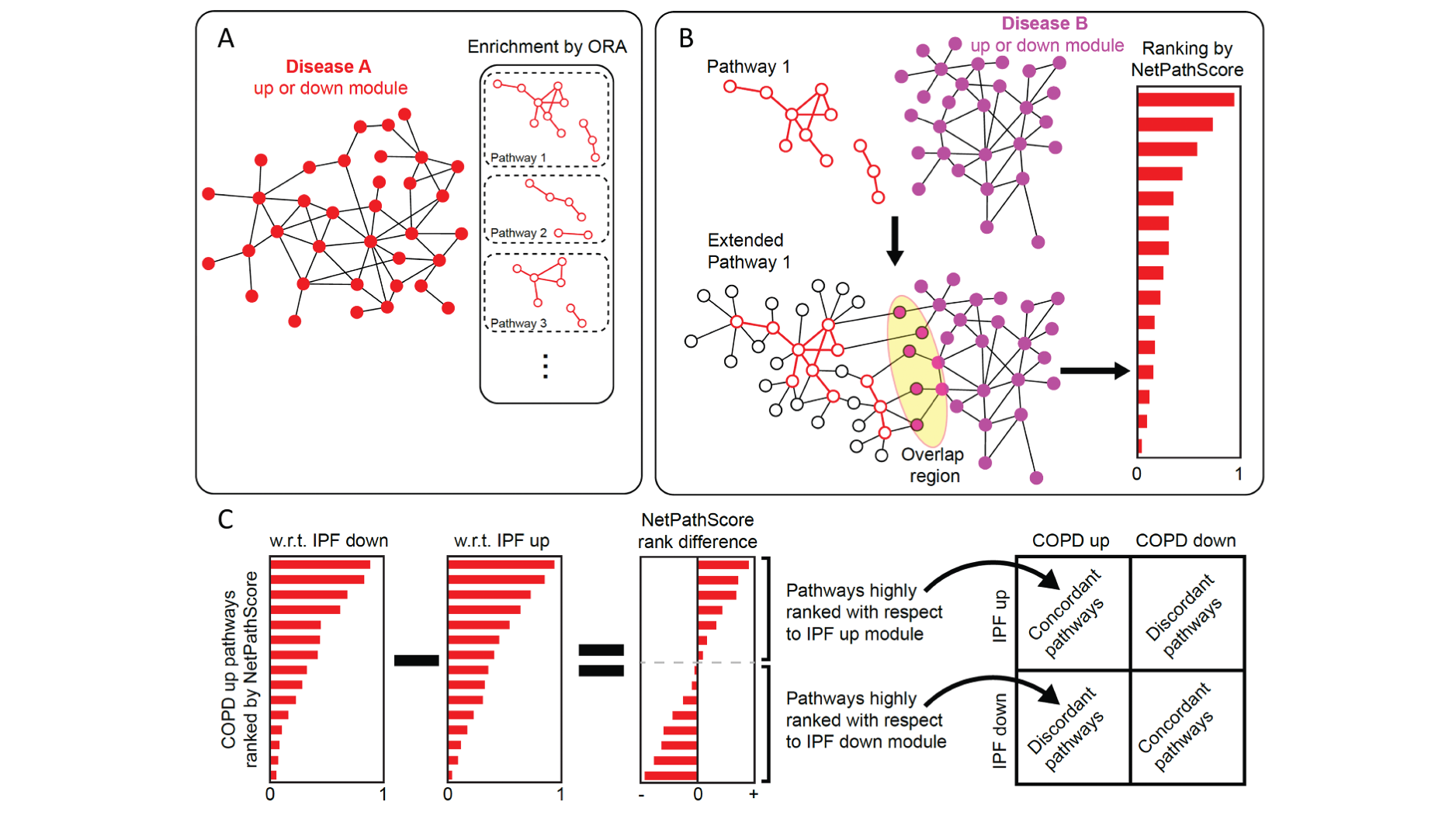
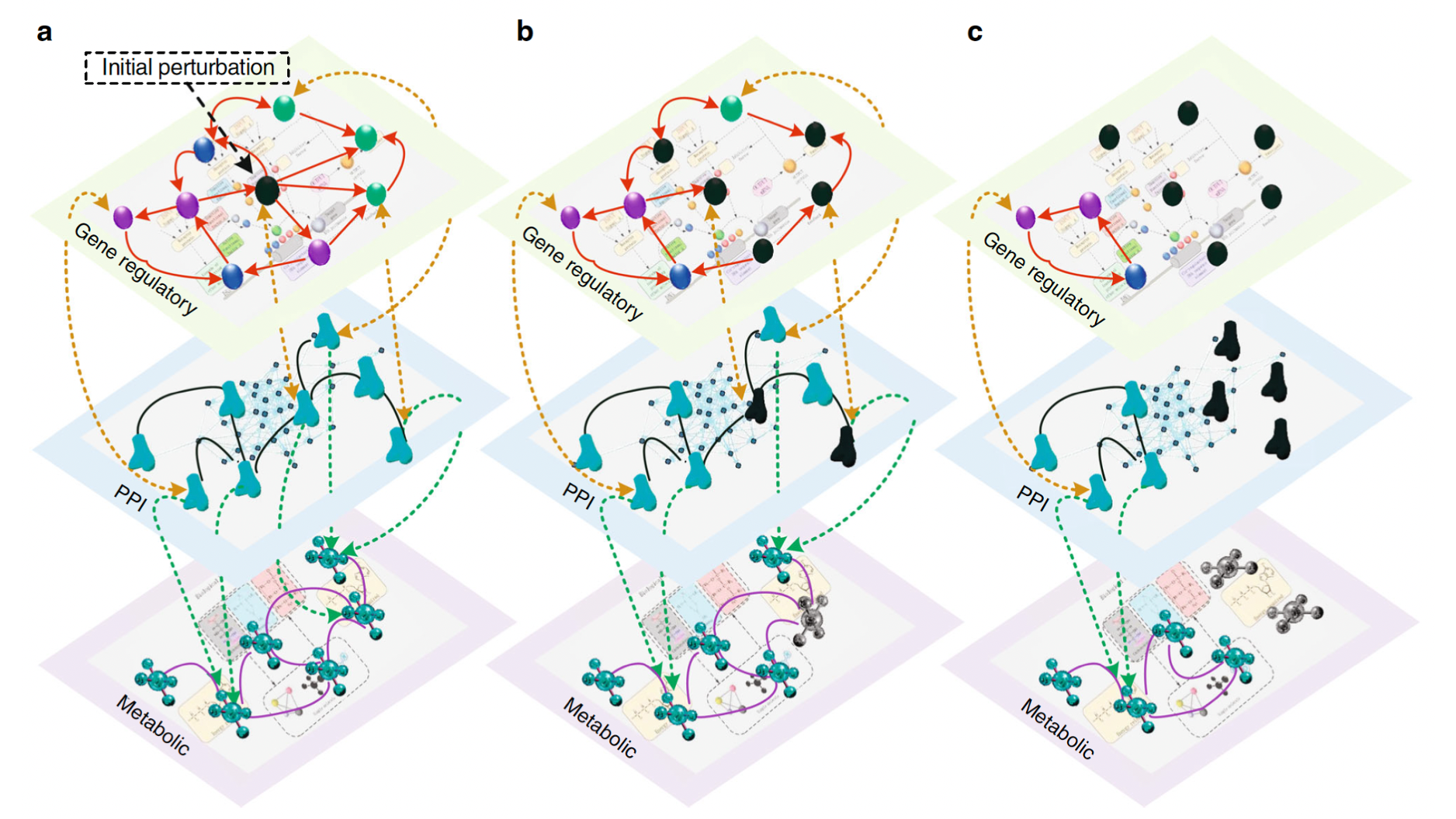
One of the primary goals of our lab is to introduce established and novel network science concepts to the biomedical domain. Our contributions in this area include (i) developing a statistical method that uses multilayer networks to model signaling transduction pathways and infer their crosstalk (Martini and Baek et al., Nucleic Acids Research), (ii) devising a network-inspired pathway ranking algorithm to identify the crosstalking pathways between COPD and IPF (Halu et al., Human Molecular Genetics), (iii) using proteomics measurements to add context-specificity to the global protein interaction network and employing network-based prioritization on this enriched interactome to predict novel regulators and potential therapeutic targets for macrophage activation (Halu et al., eLife), (iv) developing an analytical framework to model the robustness of biological systems by integrating gene regulatory, protein-protein interaction and metabolomic data in a multilayer molecular network (Liu, Maiorino and Halu et al., Nature Communications), (v) using the concept of network controllability on a pancreatic islet-specific regulatory network to determine the key pathobiological pathways in type 2 diabetes (T2D) and identified NFATC4 as the regulator of numerous putative downstream T2D candidate genes (Sharma and Halu et al., npj Systems Biology and Applications).
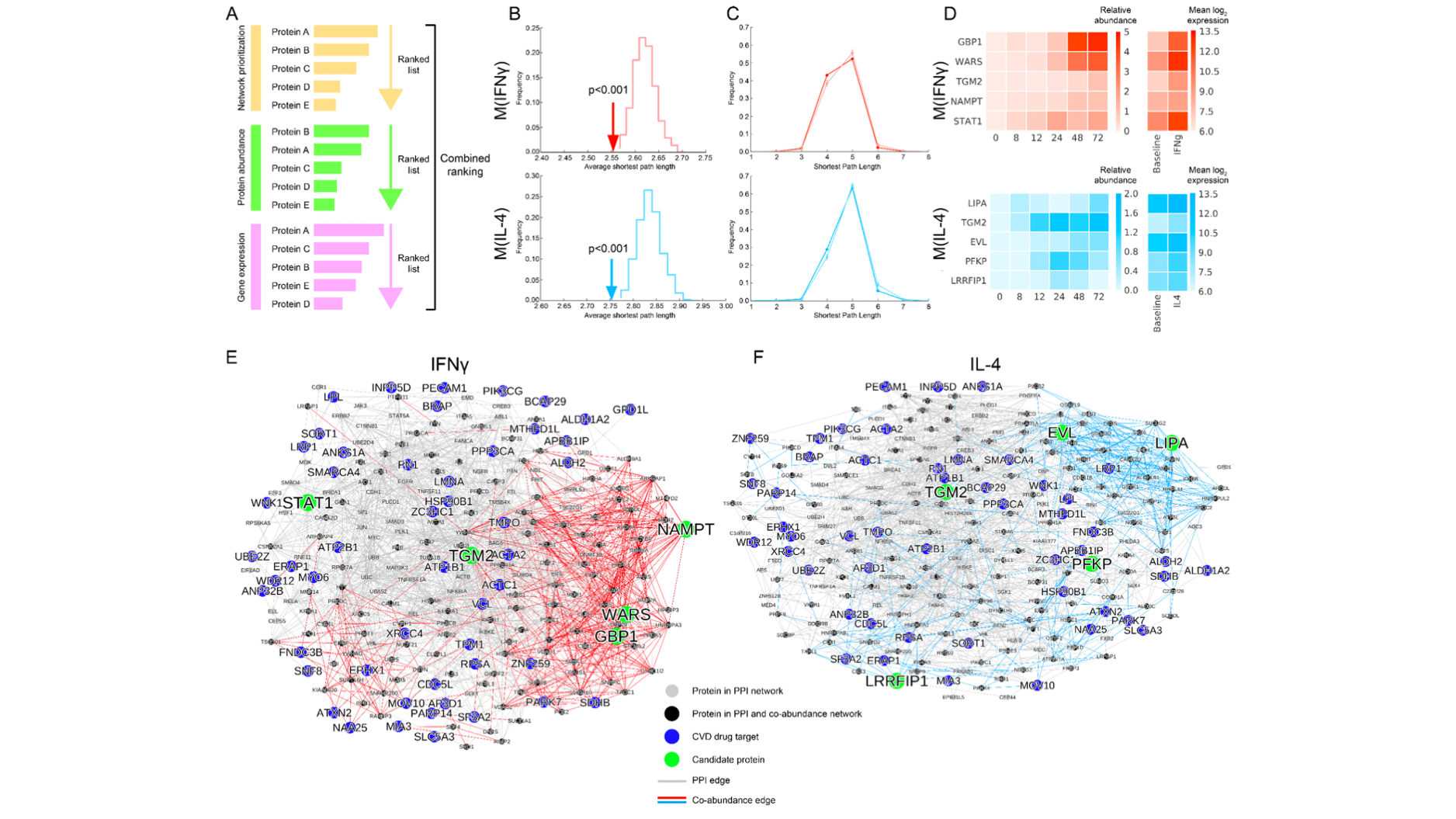
Another important part of our research is done in close collaboration with bench scientists and clinicians where we analyze multiple omic modalities and apply statistical and network-based methods to predict key regulators and pathways of complex chronic diseases. Some highlights of our contributions are (i) integrating GWAS, ChIP-seq and eQTL data to reveal the genetic overlap between vitamin D binding and asthma, autoimmune and allergic diseases in the chromosome regions 17q12-21.1 and 17q21.2 (Kilic and Halu et al., eLife), (ii) building the first spatiotemporal molecular atlas of calcific aortic valve disease (CAVD) through bioinformatics and the network analysis of transcriptomic and proteomic data, and determining the key drivers of various stages of CAVD as well as the molecules characterizing the spatial layers of the aortic valve (Schlotter and Halu et al., Circulation), (iii) integrating vesicular proteomics and transcriptomics using network-based approaches to reveal novel pathways whereby tissue extracellular vesicles modulate cardiovascular disease (Blaser et al., Circulation).
We have also contributed to personalized medicine efforts that aim to define and classify diseases by their molecular signatures. Despite their extensive use, current clinical observation-based disease taxonomies such as the International Classification of Diseases (ICD) lack the molecular depth required for precision medicine and fall short of incorporating the ever-growing amount of biomedical data. To go beyond the rigid hierarchical structures of current disease classifications and account for the phenotypic diversity and genetic heterogeneity of complex diseases, we built the multiplex network of human diseases composed of a genotypic and a phenotypic layer and used cutting-edge multiplex community detection methods to identify groups of diseases similar to each other in terms of both phenotypic and genotypic measures (Halu et al., npj Systems Biology and Applications). In addition, we were a part of a collaboration to develop an algorithm that predicts the additional categories of a disease by integrating multiple networks consisting of disease phenotypes and their molecular profiles (Zhou, Lei, Liu, Halu, et al., eBiomedicine).